Photo Credit: Singapore-MIT Alliance for Research and Technology (SMART)
Researchers from Critical Analytics for Manufacturing Personalized-Medicine (CAMP), an Interdisciplinary Research Group (IRG) at the Singapore-MIT Alliance for Research and Technology (SMART), MIT’s research enterprise in Singapore, have developed a new method for rapid and accurate detection of viral nucleic acids – a breakthrough that can be easily adapted to detect different DNA/RNA targets in viruses like the coronavirus.
While RT-qPCR is considered a gold standard for viral detection, there are limitations and they can often produce variable results. An accurate version is the digital PCR method demands a relatively long reaction time, need expensive equipment, precise temperature control, and cycling.
The new methodological development by CAMP – the RApid DIgital Crispr Approach (RADICA) – allows absolute quantification of viral nucleic acids in 40-60 minutes in an isothermal manner in a water bath, a prototypical and inexpensive laboratory equipment. The team’s research is explained in a paper titled “Digital CRISPR-based method for the rapid detection and absolute quantification of nucleic acids” published recently in the prestigious journal Biomaterials.
The RADICA method has been tested on SARS-CoV-2 synthetic DNA/RNA as well as the Epstein-Barr virus in cultured B cells and patient serum. The researchers say the method can be adapted to detect other kinds of viruses, and in other types of samples such as saliva and cell culture media. RADICA is also able to distinguish the virus from its close relatives.
“This is the first reported method of detecting nucleic acids to utilise the sensitivity of isothermal amplification and specificity of CRISPR based detection in a digital format – allowing rapid and specific amplification of DNA without the time consuming and costly need for thermal cycling,” says Dr Xiaolin Wu, Postdoctoral Associate at SMART CAMP. “RADICA offers four times faster absolute quantification compared to conventional digital PCR methods.”
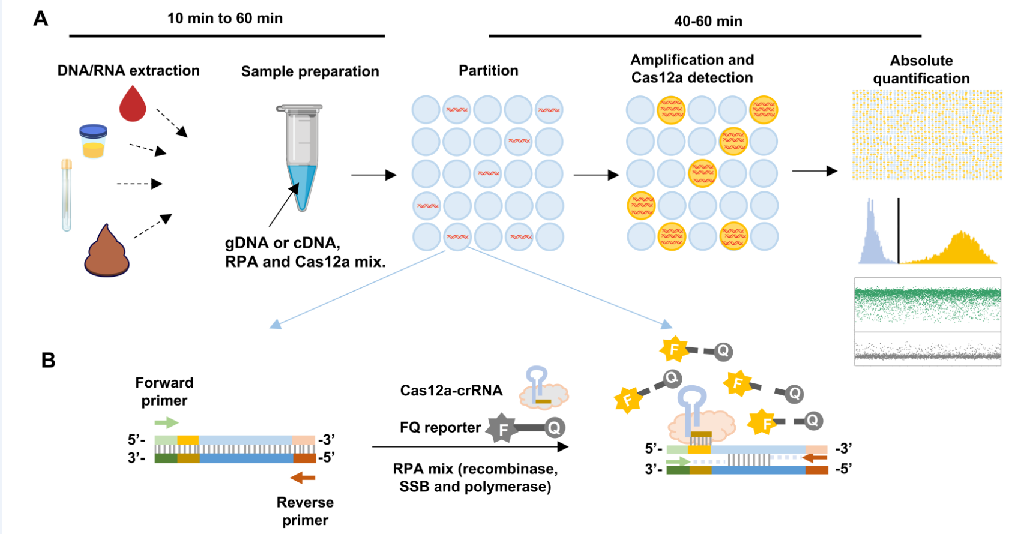
Researchers use thousands of a 15 µL DNA/RNA sample and are amplified and identified by Cas12a protein, an enzyme that can turn the target signal into a fluorescent signal. This allows absolute quantification to be achieved by counting the number of samples that have the target DNA/RNA and are lit up.
Current sterility tests need around 14 days, which is too slow for clinical needs but RADICA shortens the process into hours. RADICA also identifies how many viruses there are in the sample which can help doctors and researchers in deciding the course of treatment, as well as production and inventory management of cell therapy products. The method can also be used to detect DNA/RNA targets of different viruses and adapted to devices commonly found in hospitals and service laboratories – providing a potential new way to tackle pandemics.
The research is carried out by SMART and supported by the National Research Foundation (NRF) Singapore under its Campus for Research Excellence And Technological Enterprise (CREATE) programme.
Image Caption: “Figure shows the schematic illustration of RADICA. After DNA/RNA extraction, samples can be used for the detection and quantification of various targets. RPA-Cas12a reaction in each positive partition exponentially amplifies the target DNA, triggering more Cas12a activation and increasing the fluorescence readout. In this way, the reaction time of RADICA is reduced 4 times (from 4h to 1h compared to dPCR).”




