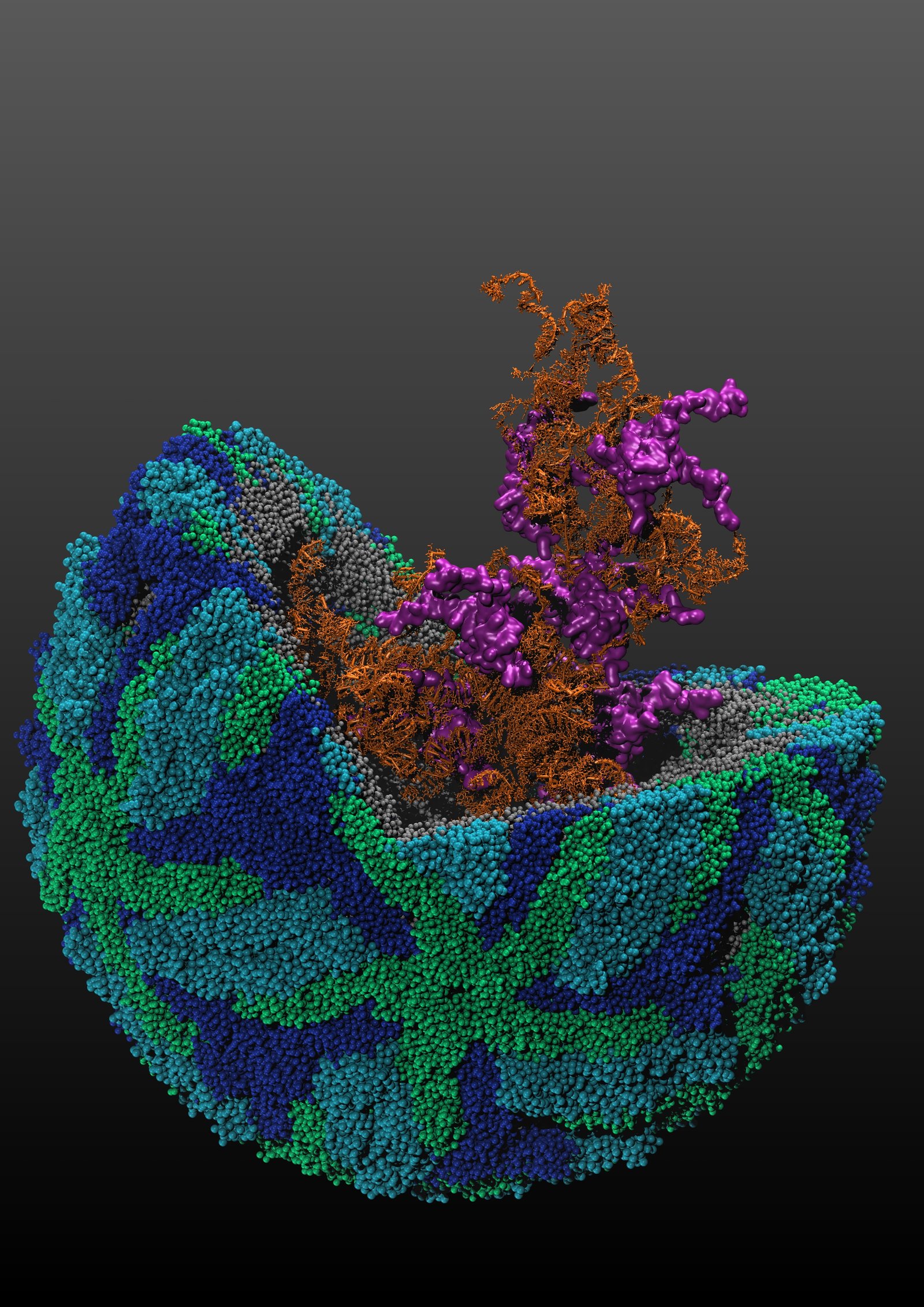
Scientists from the Agency for Science, Technology and Research’s (A*STAR) Genome Institute of Singapore (GIS), together with scientists from Duke-NUS Medical School and A*STAR’s Bioinformatics Institute (BII), have mapped out the structures of four dengue and four Zika viruses. The research focuses on understanding the genetic materials and shapes of these viruses, as well as their corresponding functions.
Consequently, understanding their biology enables scientists to design better drugs and treatments.
Mosquito-borne infections are common and deadly in tropical and subtropical regions around the world. Estimates suggest up to 700 million infections and over one million deaths are caused by a variety of mosquito-borne viruses yearly. One major class of mosquito-borne viruses are the Flaviviruses, which include the dengue and Zika viruses.
Dengue is a well-known virus which affects more than 390 million people around the world. Singapore is one of the major hotspots of dengue infection.
Every year, 2,000 to 3,000 people are affected by dengue in Singapore. Whilst most of these cases are fairly mild, dengue-associate deaths still occur each year. While the Zika virus has generally limited severity in healthy adults, there has been greater public interest due to the Zika epidemic from 2015 to 2016, and reports of adverse health effects observed in South America.
Dr Wan Yue, Principal Investigator at A*STAR’s GIS and lead author of the study, said, “One needs to understand the shape and mechanism of a lock, in order to design a key that can open it. Similarly, mapping the shapes and structures of the dengue and Zika viruses will boost understanding of their biology, and contribute significantly towards designing drug treatments targeted at these viruses.”
“Moving ahead, additional research needs to be performed to better understand how the viruses’ structural mutations reduce their effect. It will also identify the roles that different structures play during their lifecycles,” she added.
Dr Roland G. Huber, Assistant Principal Investigator at A*STAR’s BII said, “Understanding the structures of these viruses required us to develop innovative techniques to find synergies in the vast amount of data generated for this study. We developed new tools that will allow us to better understand the architecture of these and other viruses in the future. A strong team of researchers with complementary skills enabled us to succeed.”
Professor Ng Huck Hui, Executive Director at A*STAR’s GIS, said, “The collaborative study with BII and Duke-NUS unravels significant information on the genetics of the dengue and Zika viruses. This is a crucial step towards designing targeted therapeutics to tackle the diseases.”
Using structure probing, the scientists mapped the long genetic materials of four dengue and four Zika virus strains. The research studied the viruses’ activities and vital parts to identify portions of the viruses’ genetic materials that could be targeted for treatments. Through the identification of the common shapes and structures of the viruses, scientist will be able to design new drug treatments to target these areas.